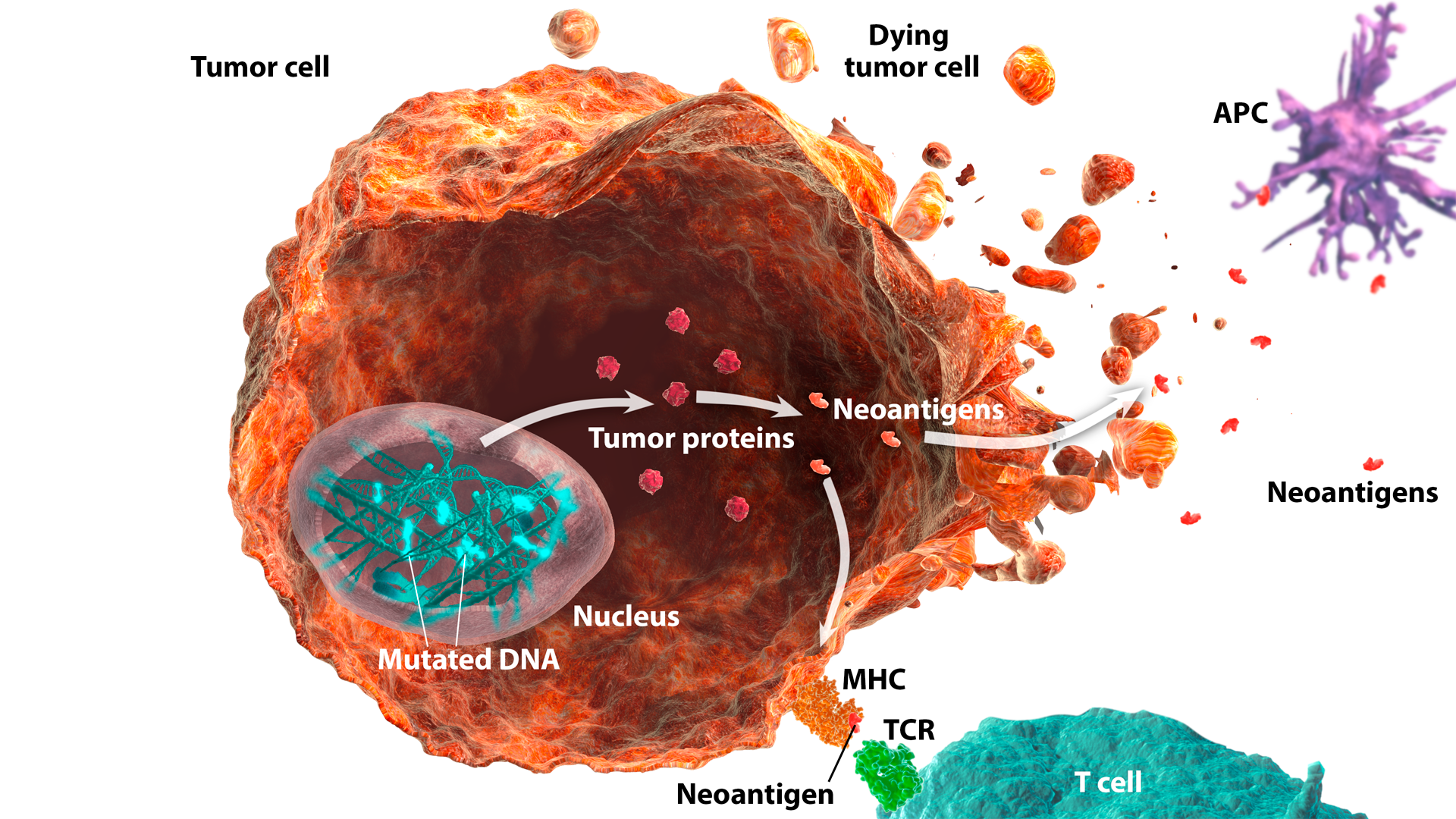
Tumor Antigens
Tumor antigens are recognized as nonself or foreign by the host immune system.1 They can initiate the adaptive immune response, a process known as immunologic priming.1,2 Several I-O biomarkers related to tumor antigens are currently under investigation:
Neoantigens
Neoantigens are newly formed antigens that have not been previously recognized by the immune system. They can arise from altered peptides formed as a result of tumor mutations or viral proteins.1,2
Neoantigens can be recognized by the immune system as nonself, and as such, can elicit an immune response.2,3 Neoantigen-specific T cells have been identified in several human cancers.4 High tumor mutation burden (TMB) and/or microsatellite instability-high (MSI-H) or deficient mismatch repair (dMMR) status may be associated with increased neoantigen production.3,5-7
Neoantigens are under investigation as a potential I-O biomarker. As immunogenic neoantigens can be challenging to identify directly, TMB may potentially be used as a surrogate to indirectly assess neoantigen load.8,9
Tumor mutation burden (TMB)
Tumor mutation burden (TMB) is defined as the collective number of somatic (acquired) mutations in the tumor genome.1,2 The number of mutations can vary across different tumor types.3,4
Tumors with high mutation burden have the potential to generate a larger number of neoantigens, making them more immunogenic.5 The increased presence of tumor-specific neoantigens leads to an increased number of tumor-infiltrating immune cells.6 High TMB has been shown to be associated with infiltration of cytotoxic T cells into the tumor microenvironment, supporting its use as a neoantigen surrogate.7-9
Distinct mechanisms of DNA mutation, such as dMMR and exposure to environmental mutagens (eg, tobacco smoke and UV light), can lead to a higher frequency of TMB.3,10
TMB is measured using next-generation sequencing (NGS), assessing either the whole genome, whole exome, or a comprehensive gene panel.1,11 The threshold for defining a high level of TMB is currently under investigation, as levels of TMB can vary across tumors.1,4
Research to investigate the potential use of TMB as an I-O biomarker is ongoing.
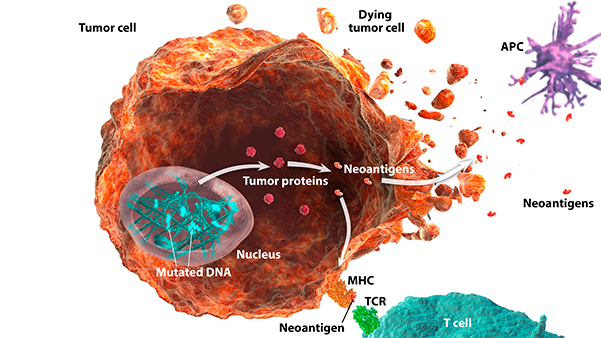
Microsatellite instability high/deficient mismatch repair
(MSI-H/dMMR)
Microsatellite instability high/deficient mismatch repair (MSI-H/dMMR) are indicators of genomic instability:
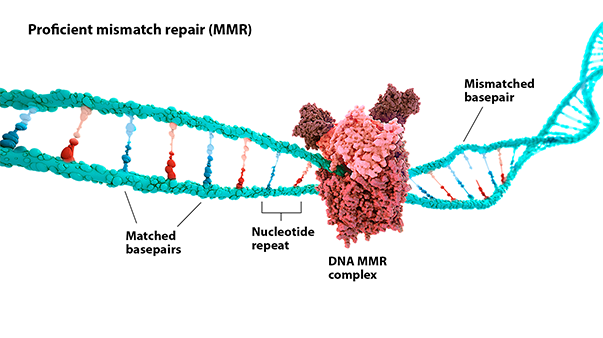
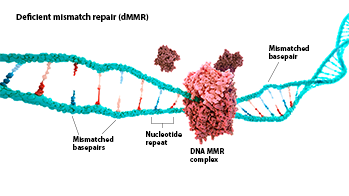
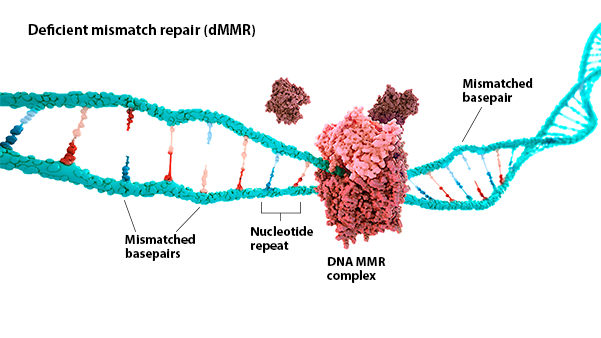
MSI-H: Microsatellite instability (MSI) is a change in the number of nucleotide repeats in DNA sequences, resulting in a different number of repeats than when the DNA was inherited.1 An MSI-H tumor has at least 2 unstable markers among 5 microsatellite markers analyzed (or ≥30% of the markers if a larger panel is used).2
dMMR: Mismatch repair (MMR) is a key DNA repair system. dMMR represents a loss of function in the MMR pathway.3
In MSI-H/dMMR tumors, more neoantigens may be produced.3,4 Neoantigens have been associated with increased T-cell activation and immune-cell infiltration of the tumor microenvironment.5,6
MSI status is most commonly detected by using polymerase chain reaction (PCR), while dMMR status is commonly detected by immunohistochemistry (IHC) for loss of expression of MMR proteins.3,7
Research to better understand the role of MSI-H/dMMR as I-O biomarkers is ongoing.
REFERENCES: Tumor antigens
1. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69-74.
REFERENCES: Neoantigens
1. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69-74. 2. Lu Y-C, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28(1):22-27. 3. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22(11):1342-1350. 4. Bobisse S, Foukas PG, Coukos G, Harari A. Neoantigen-based cancer immunotherapy. Ann Transl Med. 2016;4(14):262. 5. Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014;27(1):9-14. 6. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. 7. Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27(8):1492-1504. 8. Chabanon RM, Pedrero M, Lefebvre C, Marabelle A, Soria JC, Postel-Vinay S. Mutational landscape and sensitivity to immune checkpoint blockers. Clin Cancer Res. 2016;22(17):4309-4321. 9. Yuan J, Hegde PS, Clynes R, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer. 2016;4:3.
REFERENCES: Tumor mutation burden
1. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. 2. Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719-724. 3. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415-421. 4. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124-128. 5. Liontos M, Anastasiou I, Bamias A, Dimopoulos M-A. DNA damage, tumor mutational load and their impact on immune responses against cancer. Ann Transl Med. 2016;4(14):264. 6. Strickland KC, Howitt BE, Shukla SA, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget. 2016;7(12):13587-13598. 7. Chabanon RM, Pedrero M, Lefebvre C, Marabelle A, Soria JC, Postel-Vinay S. Mutational landscape and sensitivity to immune checkpoint blockers. Clin Cancer Res. 2016;22(17):4309-4321. 8. Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27(8):1492-1504.
REFERENCES: MSI-H/dMMR
1. National Cancer Institute. Microsatellite instability. www.cancer.gov/publications/dictionaries/cancer-terms?cdrid=285933. Accessed September 29, 2017. 2. Vilar E, Gruber SB. Microsatellite instability in colorectal cancer—the stable evidence. Nat Rev Clin Oncol. 2010;7(3):153-162. 3. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22(11):1-9. 4. Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014;27(1):9-14. 5. Giannakis M, Mu XJ, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15(4):857-865. 6. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463-1469. 7. Richman S. Deficient mismatch repair: read all about it. Int J Oncol. 2015;47(4):1189-1202.