Inflamed Tumor Markers
Inflamed tumors show evidence of immune-cell infiltration and activation in the tumor microenvironment.1 Several biomarkers exist that are reflective of an inflamed tumor microenvironment:
Programmed death ligand 1 (PD-L1)
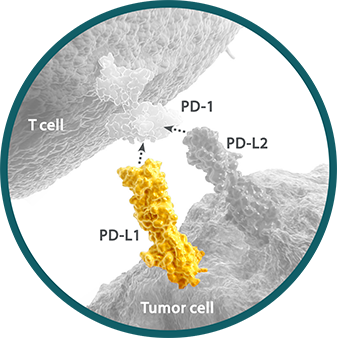
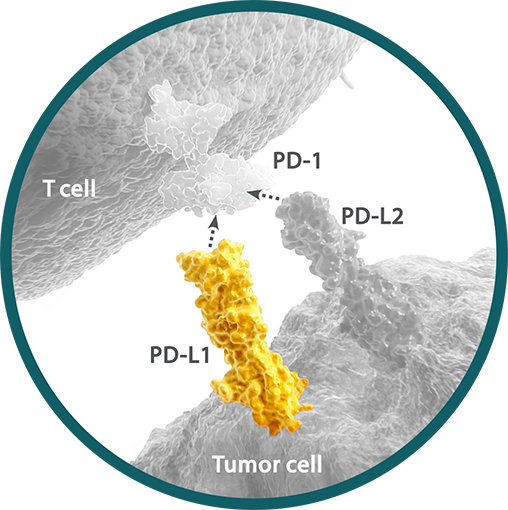
Programmed death ligand 1 (PD-L1) is a ligand for the immune checkpoint receptor programmed death receptor-1 (PD-1) expressed on the surface of cytotoxic T cells.1,2 PD-L1 expression may be present at any point along a continuum of expression. Within a tumor sample, between 0% and 100% of cells may express PD-L1.3-5
- PD-L1 expression can be determined by IHC assay and can be detected on tumor and immune cells5
- As multiple IHC assays are available, many studies have compared their analytical performance6,7
The expression of PD-L1 has implications for clinical relevance. PD-L1 expression may vary by tumor type, histology, location, and line of therapy.3,5,8,9
- Cutoff points for defining high PD-L1 expression may differ between tumor types9
Research is ongoing to better understand the role of PD-L1 as an I-O biomarker.
Programmed death ligand 2 (PD-L2)
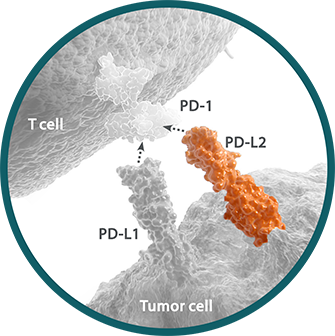
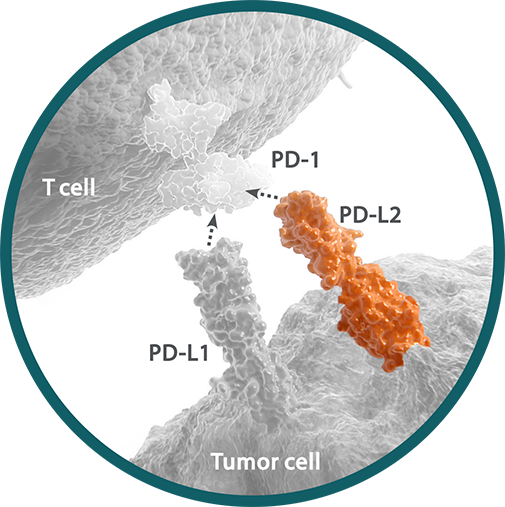
Programmed death ligand 2 (PD-L2) is a ligand for the immune checkpoint receptor PD-1 expressed on the surface of cytotoxic T cells.1,2 PD-L2 has a higher affinity than PD-L1 for PD-1, but is expressed at lower levels.3,4 PD-L2 may present at any point along a continuum of expression. Within a tumor sample, PD-L2 expression can range from absent to greater than 70%.3,5,6 Expression of PD-L2 may vary by tumor type, histology, and cell type.5,7-9 PD-L2 expression can be determined by IHC assay and detected on tumor and immune cells.3,9
PD-L2 is currently under investigation as an I-O biomarker.
Tumor-infiltrating lymphocytes (TILs)
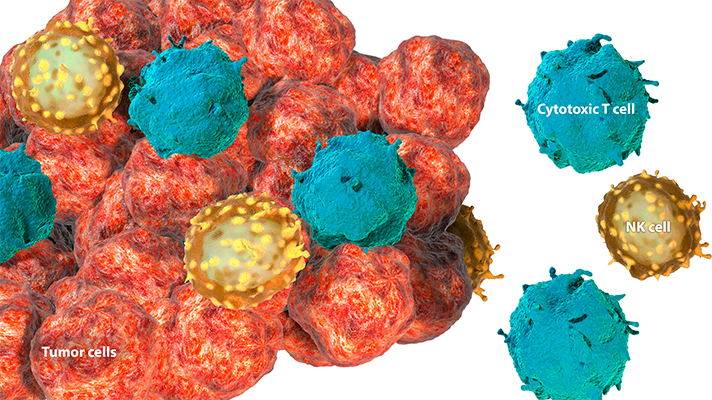
Tumor-infiltrating lymphocytes (TILs) are immune cells that enter the tumor and its microenvironment to mediate an antitumor immune response. TILs include cytotoxic T cells and natural killer (NK) cells.1 Expression of key secreted factors in the tumor microenvironment, such as chemokines and proinflammatory cytokines, can recruit these immune cells to the tumor.2
Tumors can be characterized by the extent of immune-cell infiltration. The level of TILs correlates with the degree of inflammation.3,4
The number, type, and activation state of TILs found within a tumor can be identified using techniques such as IHC and cell-sorting technology such as flow cytometry.5
TILs are currently under investigation as an I-O biomarker.
Inflammation gene signatures
Inflammation gene signatures are a specific type of gene expression profile. Gene expression profiling measures the expression of mRNA across thousands of genes.1 This can create a distinct molecular profile (or gene signature), providing a holistic view of cellular function.1,2
Reflecting the combined expression of various inflammation-specific genes, inflammation gene signatures may indicate the presence or absence of immune cells in the tumor microenvironment.2 These gene signatures can contain information on the type, amount, functional orientation, and/or location of immune cells.3
Inflammation gene signatures vary across tumor types and may be a powerful diagnostic tool.3,4 The interferon gamma (IFN-γ) gene plays a major role in the activation of the immune response.5 Consisting of IFN-γ expression along with other predefined genes, the IFN-γ signature can be used to assess the level of inflammation within the tumor microenvironment.6
To assess gene signatures, mRNA is used as an intermediary to determine the expression levels of multiple genes. Techniques include targeted mRNA expression profiling, microarrays, RNA-seq, and gene expression panels (see hypothetical example of gene expression profile below).7,8
Inflammation gene signatures are being investigated as a potential I-O biomarker.
REFERENCES: Inflamed Tumor Markers
1. Masucci GV, Cesano A, Hawtin R, et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume I - pre-analytical and analytical validation. J Immunother Cancer. 2016;4:76.
REFERENCES: PD-L1
1. Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027-1034. 2. Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261-268. 3. Gatalica Z, Snyder C, Maney T, et al. Programmed cell death 1 (PD-1) and its ligand (PD-L1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2965-2970. 4. Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti–PD-1 therapy. Clin Cancer Res. 2014;20(19):5064-5074. 5. Kerr KM, Tsao M-S, Nicholson AG, Yatabe Y, Wistuba II, Hirsch FR. Programmed death-ligand 1 immunohistochemistry in lung cancer: in what state is this art? J Thorac Oncol. 2015;10(7):985-989. 6. Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017;12(2):208-222. 7. Scheel AH, Dietel M, Heukamp LC, et al. Harmonized PD-L1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol. 2016;29(10):1165-1172. 8. Brown JA, Dorfman DM, Ma F-R, et al. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol. 2003;170(3):1257-1266. 9. Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 2016;9:5023-5039.
REFERENCES: PD-L2
1. Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261-268. 2. Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti–PD-1 therapy. Clin Cancer Res. 2014;20(19):5064-5074. 3. Rozali EN, Hato SV, Robinson BW, Lake RA, Lesterhuis WJ. Programmed death ligand 2 in cancer-induced immune suppression. Clin Dev Immunol. 2012;2012:656340. 4. Youngnak P, Kozono Y, Kozono H, et al. Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem Biophys Res Commun. 2003;307(3):672-677. 5. Calles A, Liao X, Sholl LM, et al. Expression of PD-1 and its ligands, PD-L1 and PD-L2, in smokers and never smokers with KRAS-mutant lung cancer. J Thorac Oncol. 2015;10(12):1726-1735. 6. Vanderstraeten A, Luyten C, Verbist G, Tuyaerts S, Amant F. Mapping the immunosuppressive environment in uterine tumors: implications for immunotherapy. Cancer Immunol Immunother. 2014;63(6):545-557. 7. Liu J, Liu Y, Wang W, Wang C, Che Y. Expression of immune checkpoint molecules in endometrial carcinoma. Exp Ther Med. 2015;10(5):1947-1952. 8. Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the
REFERENCES: TILs
1. Wein L, Savas P, Luen SJ, Virassamy B, Salgado R, Loi S. Clinical validity and utility of tumor-infiltrating lymphocytes in routine clinical practice for breast cancer patients: current and future directions. Front Oncol. 2017. doi:10.3389/fonc.2017.00156. 2. Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4(5):522-526. 3. Hegde PS, Karanikas V, Evers S, et al. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22(8):1865-1874. 4. Masucci GV, Cesano A, Hawtin R, et al. Validation of biomarkers to predict response to immunotherapy in cancer: Volume I - pre-analytical and analytical validation. J Immunother Cancer. 2016;4:76. 5. Ma W, Gilligan BM, Yuan J, Li T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol. 2016;9(1):47.
REFERENCES: Inflammation gene signatures
1. Walker MS, Hughes TA. Messenger RNA expression profiling using DNA microarray technology: diagnostic tool, scientific analysis or un-interpretable data? Int J Mol Med. 2008;21(1):13-17. 2. Linsley PS, Chaussabel D, Speake C. The relationship of immune cell signatures to patient survival varies within and between tumor types. PLoS One. 2015;10(9):e0138726. 3. Galon J, Angell HK, Bedognetti D, Marincola FM. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity. 2013;39(1):11-26. 4. Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014-1022. 5. Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95-109. 6. Abiko K, Matsumura N, Hamanishi J, et al. IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br J Cancer. 2015;112(9): 1501-1509. 7. Fumagalli D, Blanchet-Cohen A, Brown D, et al. Transfer of clinically relevant gene expression signatures in breast cancer: from Affymetrix microarray to Illumina RNA-Sequencing technology. BMC Genomics. 2014;15:1008. 8. Cesano A. nCounter® PanCancer Immune Profiling Panel (NanoString Technologies, Inc., Seattle, WA). J Immunother Cancer. 2015;3:42.